Effective communication of exploratory results
BUS 320
Outline
Steps in data analysis
Load the packages
Read the dataset
Glimpse the data
Rows: 20,449
Columns: 4
$ Entity <chr> "Afghanistan", "Afghanistan", …
$ Code <chr> "AFG", "AFG", "AFG", "AFG", "A…
$ Year <dbl> 1950, 1951, 1952, 1953, 1954, …
$ `Life expectancy at birth (historical)` <dbl> 27.7, 28.0, 28.4, 28.9, 29.2, …- How many rows does life_expectancy have?
Check for missing values
- Which variable has missing values? How many?
| Name | life_expectancy |
| Number of rows | 20449 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| character | 2 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| Entity | 0 | 1.00 | 4 | 59 | 0 | 256 | 0 |
| Code | 1390 | 0.93 | 3 | 8 | 0 | 237 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| Year | 0 | 1 | 1976.53 | 37.74 | 1543 | 1962.0 | 1982.0 | 2002 | 2021.0 | ▁▁▁▁▇ |
| Life expectancy at birth (historical) | 0 | 1 | 61.78 | 12.94 | 12 | 52.5 | 64.3 | 72 | 86.5 | ▁▂▅▇▅ |
Your turn
eliminate the rows that have a missing code
save the output to
life_no_missingglimpse
life_no_missinghow many rows are left?
Count obs for each Entity
- Do
Entity’s have different numbers of observations?
Randomly select 6 entities to analyze
use
set.seed()so your results will be replicablesave to
entities
Display entities
Prepare data for plotting
extract only the rows for the 6 entities
rename the last column to expectancy
drop the variable
Codesave to object
life_df
glimpse life_df
Find the min and max year
- for your entities find the min and max year
Plot Year vs expectancy for each entity
use
ggplot2to create a line plot for each entity withYearon the x-axis andexpectancyon the y-axisadd points to the plot
format the y-axis to display the values with the suffix “years”
add a title, “Life Expectancy, 1876 to 2021”, min year to max year
-
change the color using scale_color_
- look at the help for ?scale_color_viridis_d` to see how to change the color palette
life_df |>
ggplot(aes(x = Year, y = expectancy, color = Entity)) +
geom_line() +
geom_point()+
scale_y_continuous(labels = number_format(suffix = " years")) +
labs(x = NULL, y = NULL, color = NULL, title = "Life Expectancy, 1876 to 2021", caption = "Source Our World in Data (https://ourworldindata.org/life-expectancy)") +
scale_color_viridis_d(option = "plasma", begin = 0, end = 0.8)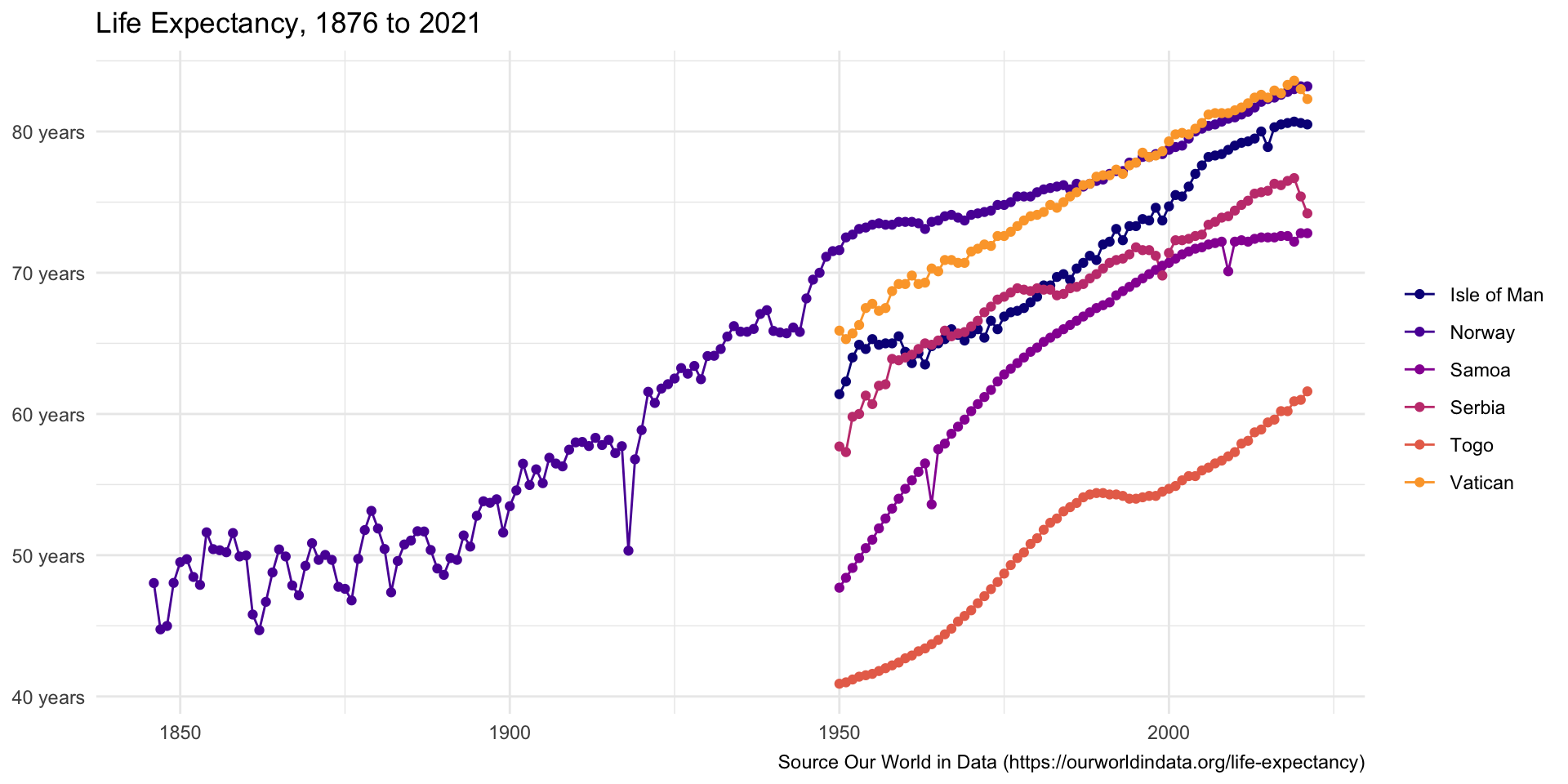
Assign the plot to an object p_life_df
p_life_df <- life_df |>
ggplot(aes(x = Year, y = expectancy, color = Entity)) +
geom_line() +
geom_point()+
scale_y_continuous(labels = number_format(suffix = " years")) +
labs(x = NULL, y = NULL, color = NULL, title = "Life Expectancy, 1876 to 2021", caption = "Source Our World in Data (https://ourworldindata.org/life-expectancy)") +
scale_color_viridis_d(option = "plasma", begin = 0, end = 0.8)Create a plotly object
Add text the plot
- add a text annotation to the plot
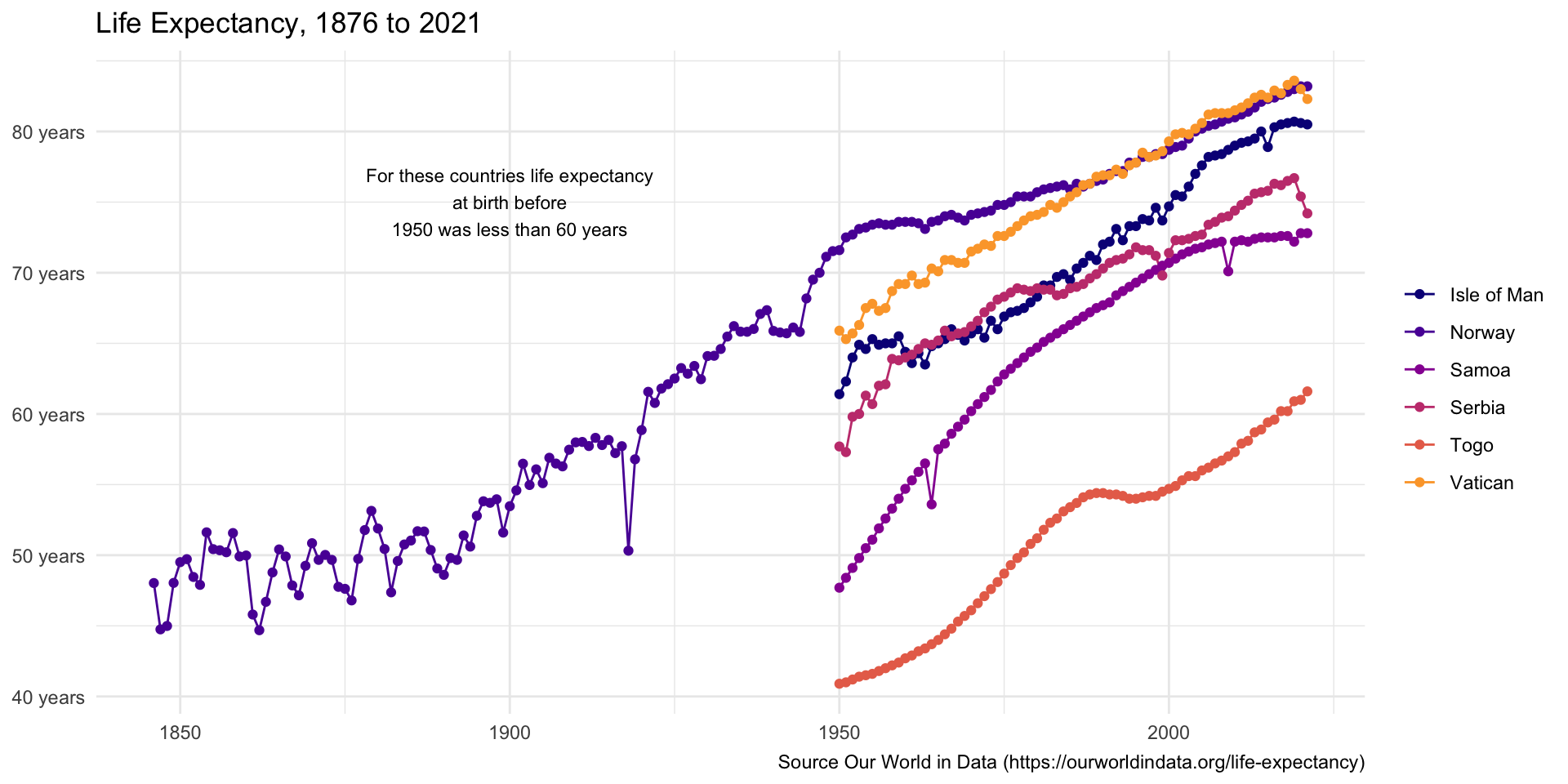
Quiz
- 4 Multiple choice questions
- 3 Numeric questions
- Quiz instructions will have a dataset that you will read in similar to
life_expectancy
